
Marcelo Mollinari, PhD
Research Assistant Professor | NCSU Bioinformatics Research Center | NCSU Department of Horticultural

Research Assistant Professor | NCSU Bioinformatics Research Center | NCSU Department of Horticultural
Welcome to my website! I am Dr. Marcelo Mollinari, a Research Assistant Professor at North Carolina State University, affiliated with both the Bioinformatics Research Center and the Department of Horticultural Science. I specialize in bioinformatics and plant genetics, leading research in diploid and polyploid breeding systems by developing unique algorithms that analyze interconnected families and translate genomic data into actionable insights. I collaborate closely with breeders of various crop species, including potatoes, sweetpotatoes, berries, and sugarcane to improve crop productivity, resilience, and sustainability. My interest lies in bridging the gap between data science and agricultural innovation, ultimately providing genetic analysis tools that support discovery and practical breeding solutions for researchers and breeders.
My research interests span a wide range of topics in bioinformatics and plant genomics, with a focus on genetic analysis in outcrossing diploid and polyploid species. Here’s a brief overview of my work:
These are some software tools I have maintained and contributed to:
SIMpoly This is a new addition to my package suite - a simple tool designed to simulate interconnected populations for testing mappoly2. I have no plans to publish or submit it to CRAN, but I’m making it available in case others find it useful.
MAPpoly2 is an advanced R package designed for constructing genetic maps in interconnected full-sib autopolyploid families. This enhanced version prioritizes user-friendliness and accessibility, with improvements geared toward seamless integration with tools like R Shiny to provide an intuitive interface for polyploid genetic mapping. The package supports ploidy levels of 2, 4, and 6, allowing for any combination of these configurations, making it highly versatile for diverse research needs.
A standout feature of MAPpoly2 is its significantly enhanced performance, achieved through the integration of computationally intensive code implemented in C++. This optimization enables the efficient analysis of large and complex datasets, addressing a common bottleneck in genetic mapping workflows.
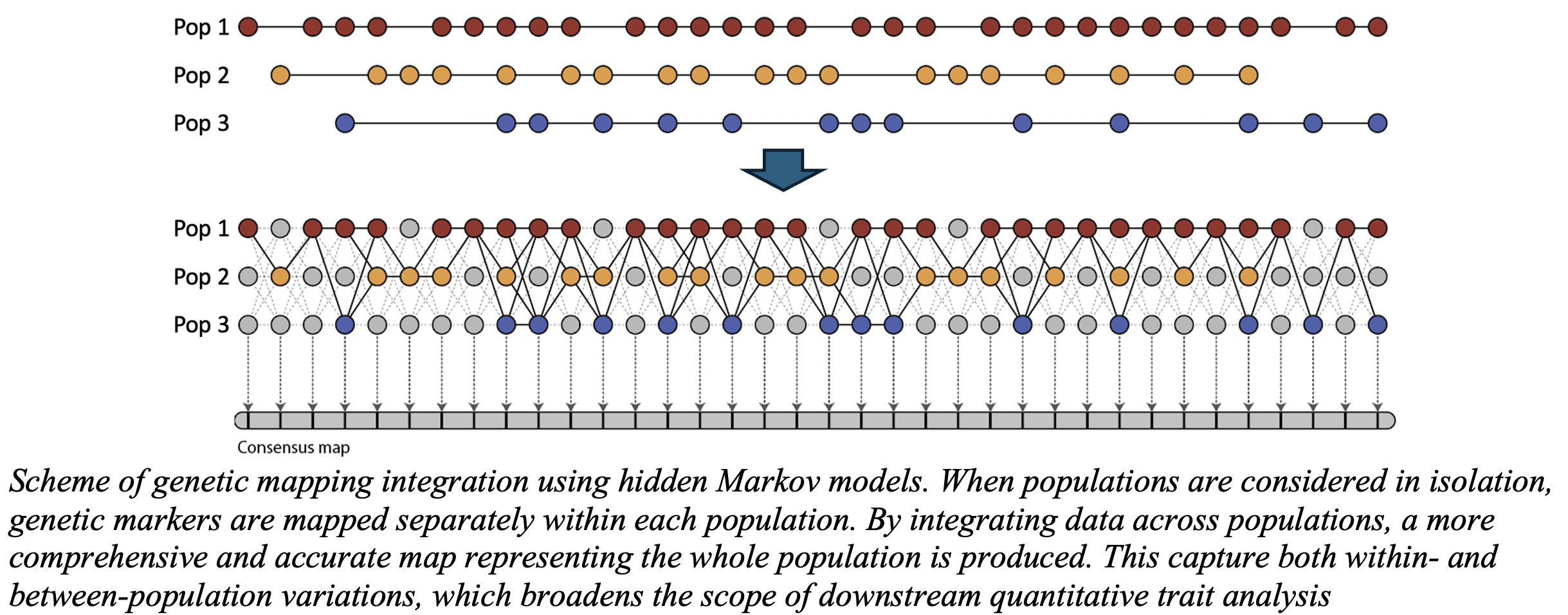
The package employs a robust Hidden Markov Model (HMM) framework to construct individual genetic maps for each parent. These maps are then combined into a comprehensive joint map, which is recomputed to incorporate any remaining markers, ensuring high accuracy and resolution in the final results. These innovations make MAPpoly2 an invaluable tool for researchers working with polyploid species and large-scale genomic datasets.

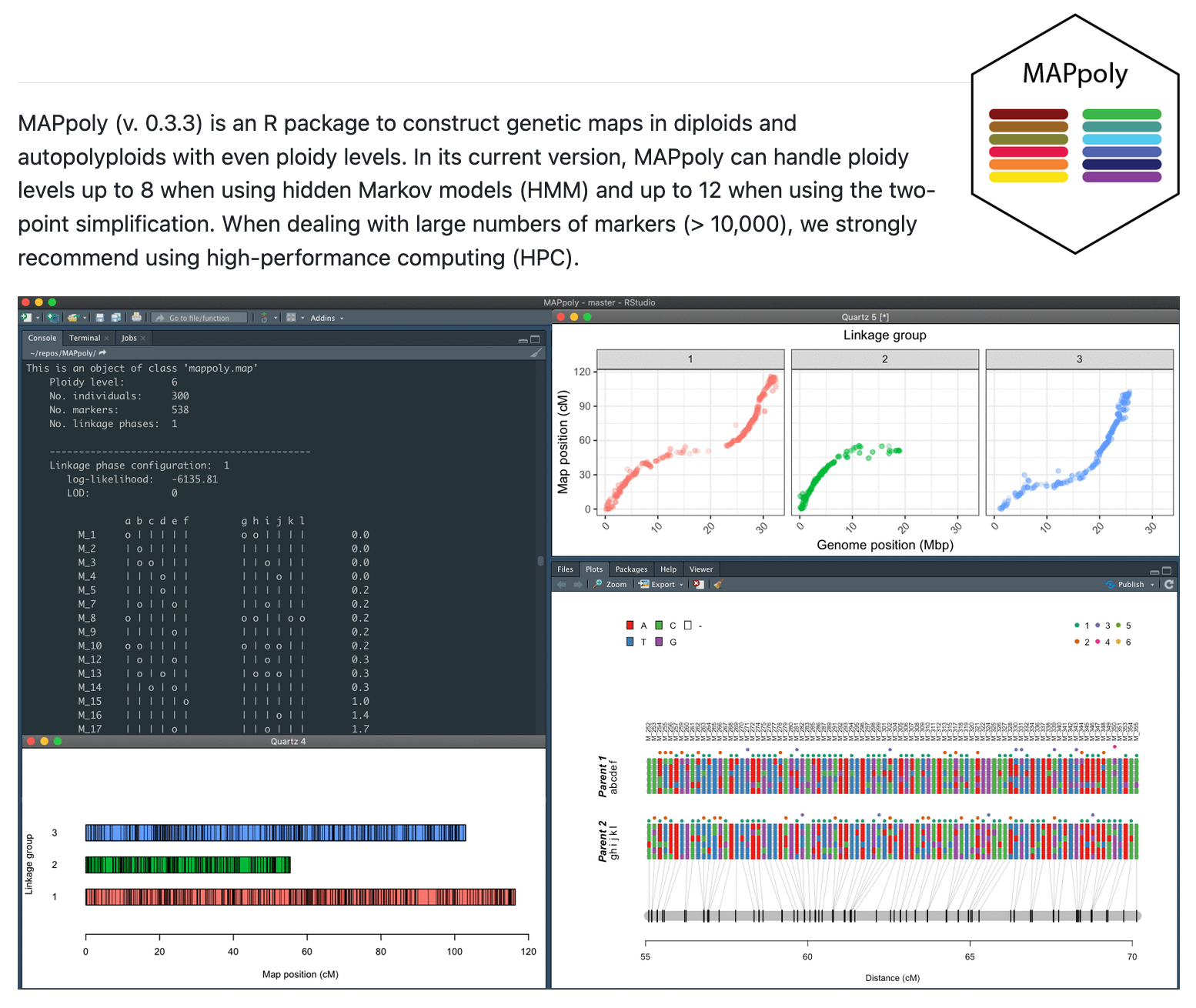
MAPpoly is a tool for genetic mapping in diploids and autopolyploids, providing statistical models...
VIEWpoly offers a powerful graphical user interface designed for integrating, visualizing, and exploring results from linkage and quantitative trait loci (QTL) analyses, along with genomic data, specifically tailored for autopolyploid and diploid species. This interactive application allows users to seamlessly upload and combine various data sources, including gene annotations and alignment files, to facilitate candidate gene exploration in an integrated genome browser.
VIEWpoly supports input from multiple tools beyond MAPpoly, including polymapR, diaQTL, QTLpoly, and polyqtlR, making it a versatile solution for researchers working with complex polyploid genetics. By enabling the integration of diverse datasets, VIEWpoly provides an interactive platform for comprehensive analysis and discovery in polyploid research.

QTLpoly The R package QTLpoly is software to map quantitative trait loci (QTL) in full-sib families of outcrossing autopolyploid species based on a random-effect multiple QTL model (Pereira et al. 2020). Variance components associated with putative QTL are tested using score statistics (Qu et al. 2013), and final models are fitted using residual maximum likelihood (REML, adapted from the R package sommer).
OneMap is a software package designed for constructing genetic maps in experimental crosses, including full-sib, RILs, F2, and backcross populations. Originally developed by Gabriel R. A. Margarido, myself (Marcelo Mollinari), and A. Augusto F. Garcia, the project later welcomed contributions from Rodrigo R. Amadeu, Cristiane H. Taniguti, and Getulio C. Ferreira. OneMap has been available on CRAN for several years, with its most recent stable version released in 2020. The CRAN version is recommended for most users, while the development version—featuring ongoing improvements and experimental functions—is hosted on GitHub. A major milestone for OneMap was the integration of support for sequencing-based markers, such as those from Illumina and GBS technologies, allowing users to analyze modern genomic datasets effectively. The package provides a comprehensive set of functions for building linkage maps, with some specialized functions running internally to optimize analysis workflows.
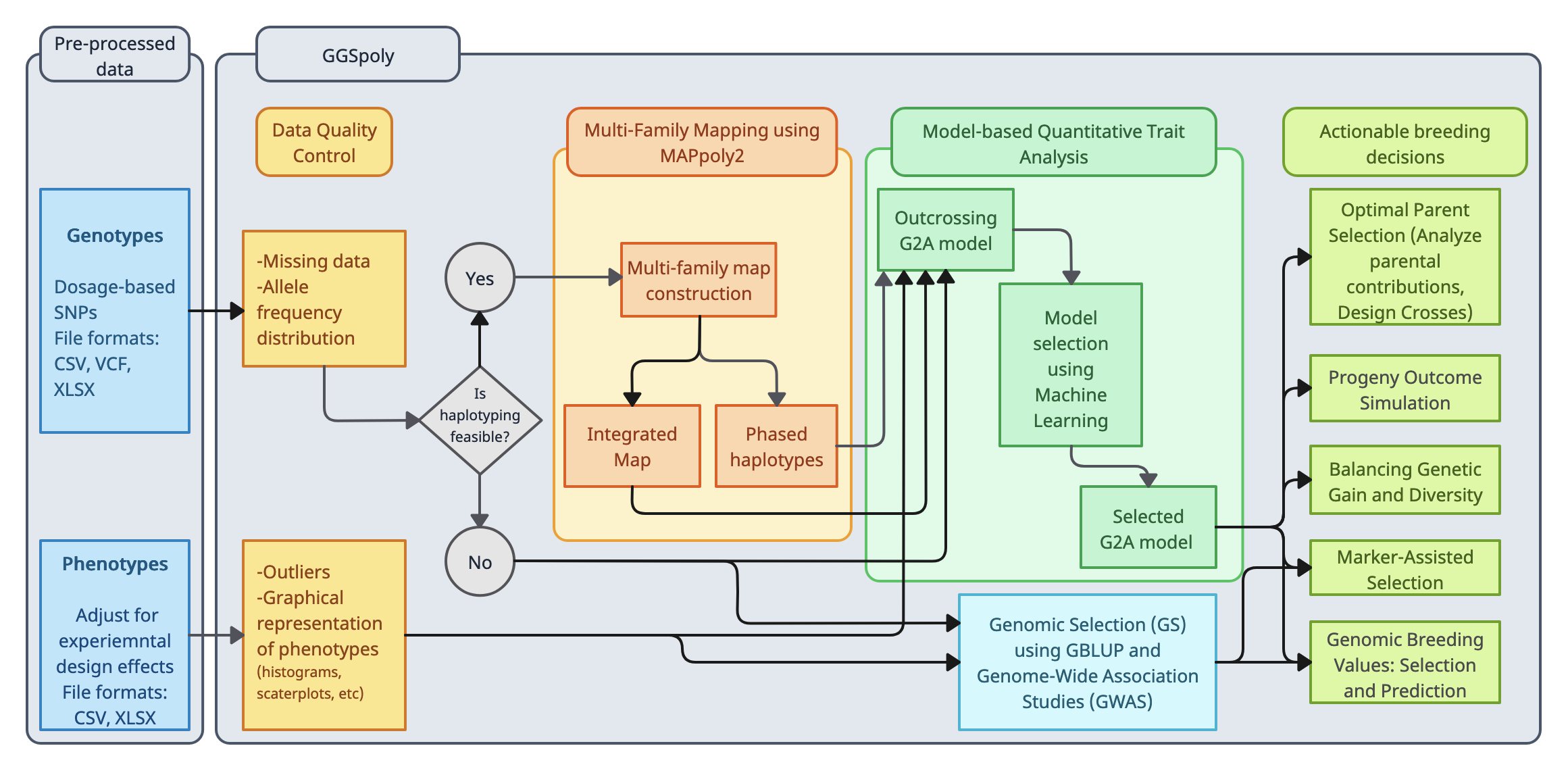
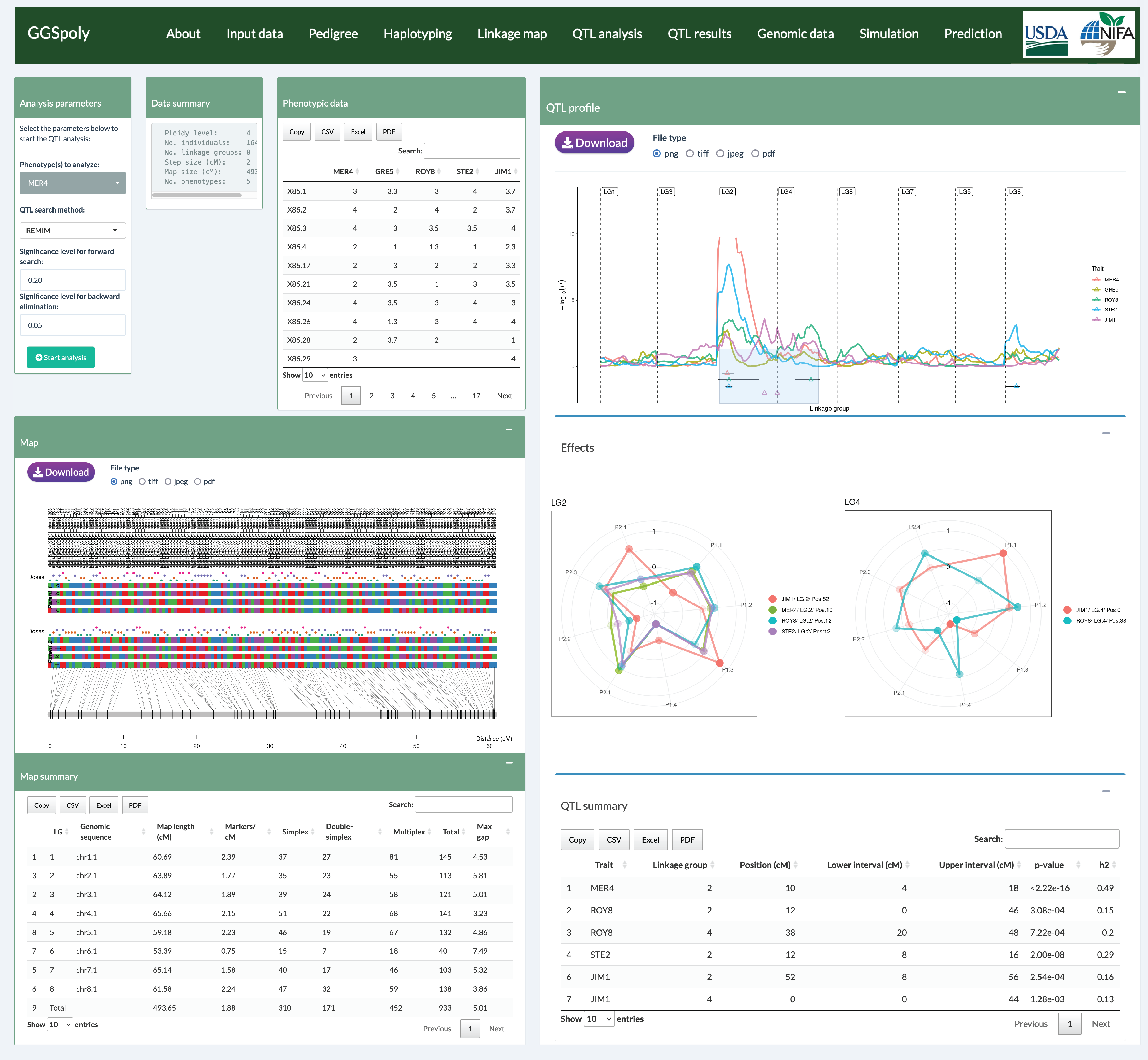
GGSpoly is a web-based tool for genomic data analysis in diploid and
polyploid species. It provides an intuitive interface for breeders and researchers to
upload, analyze, and visualize genomic data. The platform supports various genetic analysis
tasks, including haplotype inference, QTL mapping, and consensus map construction. GGSpoly
aims to bridge the gap between complex genetic models and practical breeding applications,
making advanced genomic tools accessible to a broader audience.
Here are slides from some of the talks, workshops, and lectures I've given over the years:
Explore additional scripts and resources related to my research and software development: